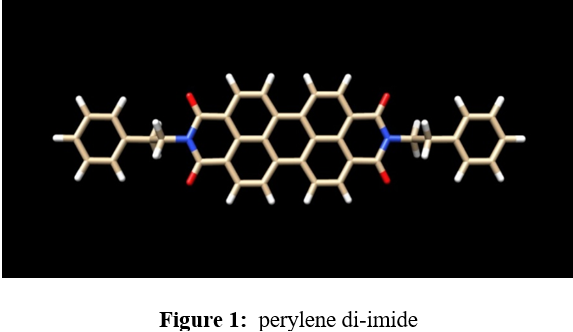
Study of perylene Di-imide as a Binder of a Human Telomeric Tetraplex DNA by Molecular Docking Simulation
Mishra V1*, Tiwari R2
DOI:https://doi.org/10.61343/jcm.v2i01.49
1* Vandana Mishra, Department of Physics, DDU Gorakhpur University, Gorakhpur, U P, India.
2 Rakesh Kumar Tiwari, Department of Physics, DDU Gorakhpur University, Gorakhpur, U P, India.
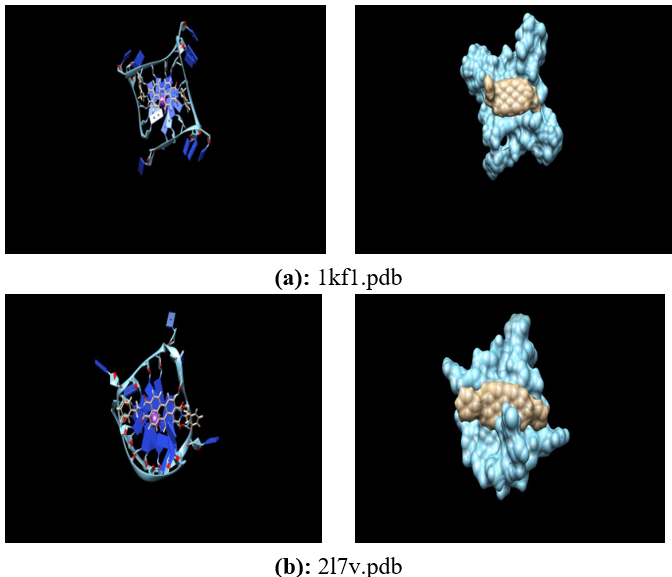
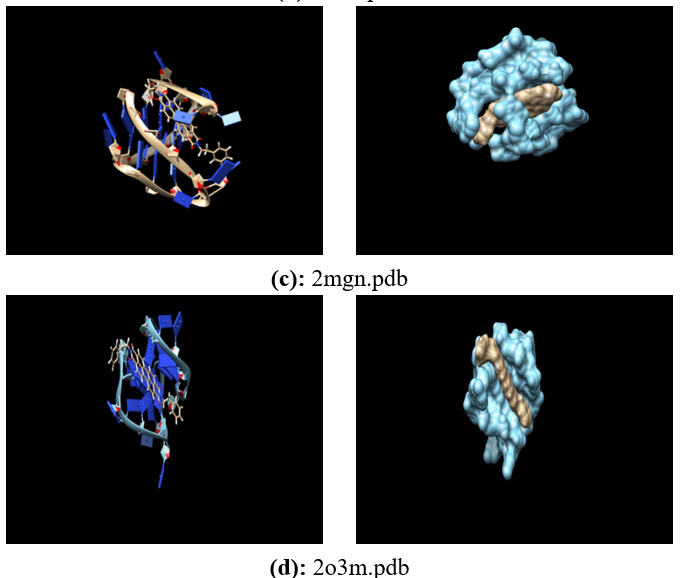
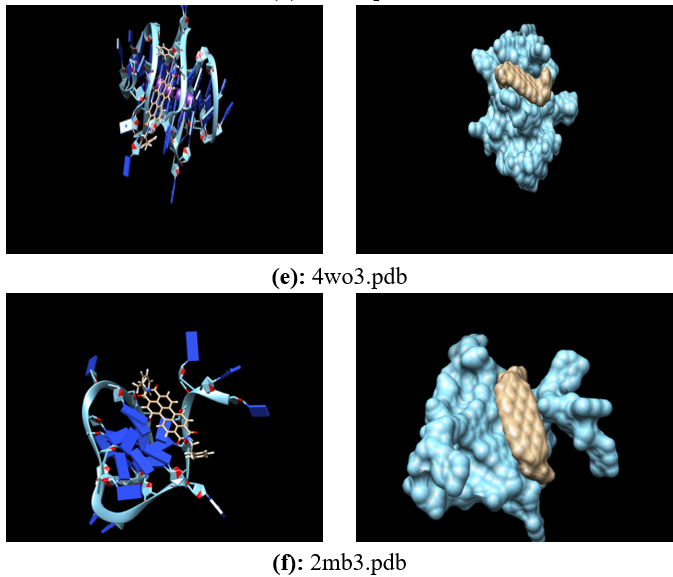
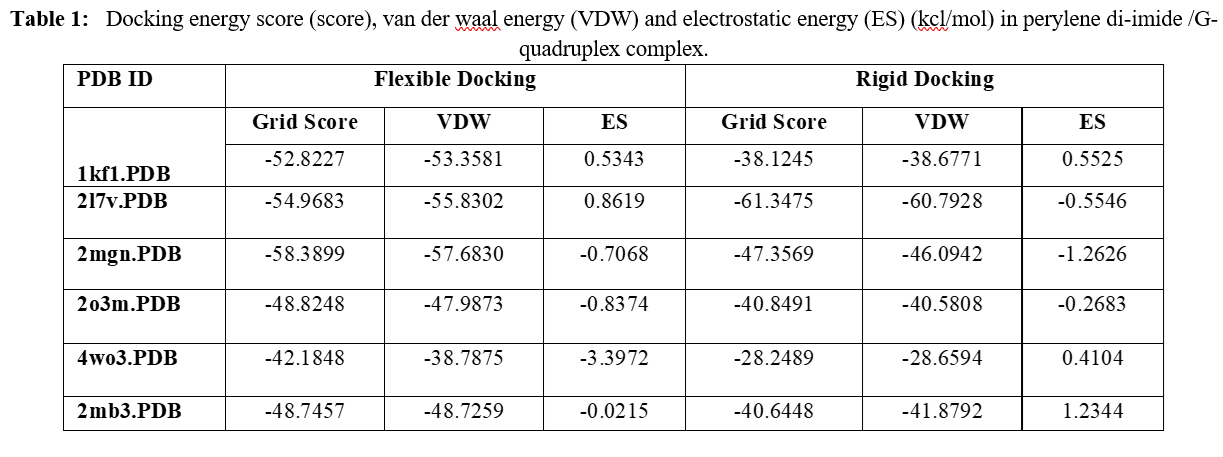
Human telomeres are consisting of d(TTAGGG) repeats involved in the formation of tetraplex DNA structures. Ligands stabilizing these tetraplex DNA structures are potential inhibitors of the cancer cell-associated enzyme telomerase. In human cells, telomerase adds multiple copies of the 5’-GGTTAG-3’ motif to the end of the G-strand of the telomere and in the majority of tumour cells it results over-expressed. Several structural studies have revealed a diversity of topologies for telomeric tetraplexes, which are sensitive to the nature of the cations present, to the flanking sequences, and probably also to concentration, as confirmed by the different conformations deposited in the Protein Data Bank (PDB). The existence of different polymorphism in the DNA quadruplex and the absence of a uniquely precise binding site give rise to a better understanding of mechanism by verifying with docking methodology approach. To target this, we have selected six different experimental models of the human telomeric sequence d[AG3(T2AG3)3] with the ligands (the perylene di-imide). We simulated out molecular docking of binding of perylene di-imide to a selected G-quadruplex using dock 6.9 to examine the or loop binding mode of perylene di-imide.
The simulation provides the two highest rank docking poses of perylene di-imide i.e. with external strand and groove binding mode; the role different ligand binding is described as external striking on the terminal guanine tetrad and the groove binding. which may be further considered for the study of therapeutic properties of ligand.
Keywords: DNA Duplex, Docking, telomeric quadruplex and molecular interaction
| Corresponding Author | How to Cite this Article | To Browse |
|---|---|---|
| , , Department of Physics, DDU Gorakhpur University, Gorakhpur, U P, India. Email: |
Mishra V, Tiwari R, Study of perylene Di-imide as a Binder of a Human Telomeric Tetraplex DNA by Molecular Docking Simulation. J.Con.Ma. 2024;2(1):20-23. Available From |


 ©
©